Coomassie Protein Assay Reagent Sds
The Bradford protein assay was adult by Marion 1000. Bradford in 1976.[one] It is a quick and accurate[two] spectroscopic analytical procedure used to measure the concentration of protein in a solution. The reaction is dependent on the amino acid composition of the measured proteins.
Principle [edit]
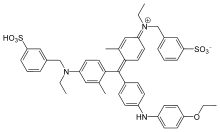
Effigy 1. Coomassie bright blue G-250, the binding dye for the Bradford Method

Color reaction of protein and Bradford reagent
The Bradford assay, a colorimetric protein assay, is based on an absorbance shift of the dye Coomassie bright blue G-250. The Coomassie brilliant bluish G-250 dye exists in three forms: anionic (blueish), neutral (green), and cationic (ruddy).[iii] Nether acidic conditions, the red class of the dye is converted into its bluish form, binding to the protein being assayed. If there'due south no protein to bind, and so the solution will remain dark-brown. The dye forms a potent, noncovalent complex with the protein'due south carboxyl group by van der Waals forcefulness and amino group through electrostatic interactions.[1] During the germination of this complex, the crimson form of Coomassie dye beginning donates its free electron to the ionizable groups on the poly peptide, which causes a disruption of the protein's native state, consequently exposing its hydrophobic pockets. These pockets in the protein's third structure demark non-covalently to the not-polar region of the dye via the first bond interaction (van der Waals forces) which position the positive amine groups in proximity with the negative charge of the dye. The bond is farther strengthened past the second bond interaction between the two, the ionic interaction. When the dye binds to the poly peptide, information technology causes a shift from 465 nm to 595 nm, which is why the absorbance readings are taken at 595 nm.[4]
The cationic (unbound) grade is light-green / ruddy and has an absorption spectrum maximum historically held to be at 465 nm. The anionic bound class of the dye which is held together past hydrophobic and ionic interactions, has an absorption spectrum maximum historically held to be at 595 nm.[5] The increment of absorbance at 595 nm is proportional to the amount of bound dye, and thus to the corporeality (concentration) of poly peptide present in the sample.[6]
Different other protein assays, the Bradford poly peptide assay is less susceptible to interference by various chemical compounds such every bit sodium, potassium or even carbohydrates like sucrose, that may be present in protein samples.[ii] An exception of note is elevated concentrations of detergent. Sodium dodecyl sulfate (SDS), a mutual detergent, may be found in protein extracts because information technology is used to lyse cells by disrupting the membrane lipid bilayer and to denature proteins for SDS-PAGE. While other detergents interfere with the assay at high concentration, the interference caused by SDS is of 2 different modes, and each occurs at a different concentration. When SDS concentrations are below critical micelle concentration (known as CMC, 0.00333%Westward/Five to 0.0667%) in a Coomassie dye solution, the detergent tends to bind strongly with the protein, inhibiting the protein bounden sites for the dye reagent. This tin cause underestimations of poly peptide concentration in solution. When SDS concentrations are above CMC, the detergent associates strongly with the greenish form of the Coomassie dye, causing the equilibrium to shift, thereby producing more of the blue form. This causes an increment in the absorbance at 595 nm contained of protein presence.[half dozen]
Other interference may come up from the buffer used when preparing the protein sample. A high concentration of buffer volition cause an overestimated protein concentration due to depletion of free protons from the solution by conjugate base from the buffer. This will non be a problem if a low concentration of poly peptide (after the buffer) is used.[six]
In order to mensurate the absorbance of a colorless chemical compound a Bradford assay must be performed. Some colorless compounds such as proteins tin can be quantified at an Optical Density of 280 nm due to the presence of effluvious rings such as Tryptophan, Tyrosine and Phenylalanine but if none of these amino acids are present then the absorption cannot be measured at 280 nm.[7]
Advantages [edit]
Many protein-containing solutions take the highest absorption at 280 nm in the spectrophotometer, the UV range. This requires spectrophotometers capable of measuring in the UV range, which many cannot. Additionally, the assimilation maxima at 280 nm requires that proteins contain aromatic amino acids such as tyrosine (Y), phenylalanine (F) and/or tryptophan (West). Not all proteins contain these amino acids, a fact which will skew the concentration measurements. If nucleic acids are present in the sample, they would also absorb lite at 280 nm, skewing the results further. By using the Bradford protein assay, one can avoid all of these complications by simply mixing the protein samples with the Coomassie brilliant blueish G-250 dye (Bradford reagent) and measuring their absorbances at 595 nm, which is in the visible range.[8]
The procedure for Bradford poly peptide analysis is very easy and simple to follow. It is washed in i pace where the Bradford reagent is added to a test tube forth with the sample. Subsequently mixing well, the mixture about immediately changes to a blue color. When the dye binds to the proteins through a process that takes near ii minutes, a change in the absorption maximum of the dye from 465 nm to 595 nm in acidic solutions occurs.[2] This dye creates strong noncovalent bonds with the proteins, via electrostatic interactions with the amino and carboxyl groups, too as Van Der Waals interactions. Merely the molecules that bind to the proteins in solution exhibit this change in absorption, which eliminates the business organisation that unbound molecules of the dye might contribute to the experimentally obtained assimilation reading. This process is more benign since it is less pricey than other methods, easy to utilize, and has high sensitivity of the dye for protein.[nine]
Later on 5 minutes of incubation, the absorbance can be read at 595 nm using a spectrophotometer; an easily accessible motorcar.
This assay is i of the fastest assays performed on proteins.[10] The full time information technology takes to set up and complete the assay is under 30 minutes.[11] The entire experiment is washed at room temperature.
The Bradford poly peptide assay can measure protein quantities as piddling as 1 to 20 μg.[12] Information technology is an extremely sensitive technique.
The dye reagent is a stable set up to use product prepared in phosphoric acid. It can remain at room temperature for up to two weeks before it starts to degrade.
Protein samples commonly incorporate salts, solvents, buffers, preservatives, reducing agents and metallic chelating agents. These molecules are frequently used for solubilizing and stabilizing proteins. Other protein assay like BCA and Lowry are ineffective because molecules like reducing agents interfere with the analysis.[thirteen] Using Bradford can exist advantageous confronting these molecules because they are compatible to each other and volition non interfere.[14]
The linear graph acquired from the analysis (absorbance versus protein concentration in μg/mL) tin can be easily extrapolated to make up one's mind the concentration of proteins by using the gradient of the line.
It is a sensitive technique. It is also very unproblematic: measuring the OD at 595 nm later 5 minutes of incubation. This method can also make apply of a Vis spectrophotometer.[fifteen]
Disadvantages [edit]
The Bradford assay is linear over a short range, typically from 0 µg/mL to 2000 µg/mL, ofttimes making dilutions of a sample necessary before analysis. In making these dilutions, fault in one dilution is compounded in further dilutions resulting in a linear relationship that may non always be accurate.
Basic weather and detergents, such equally SDS, tin interfere with the dye's ability to bind to the protein through its side chains.[10] However, there are some deterd protein, information technology is possible that the concentration measured will be inaccurate.
The reagents in this method tend to stain the test tubes. Aforementioned exam tubes cannot exist used since the stain would affect the absorbance reading. This method is also time sensitive. When more than than one solution is tested, it is important to make certain every sample is incubated for the same amount of time for accurate comparison.[16]
It is also inhibited by the presence of detergents, although this problem can exist alleviated by the addition of cyclodextrins to the assay mixture.[17]
Much of the non-linearity stems from the equilibrium between two different forms of the dye which is perturbed by adding the protein. The Bradford assay linearizes by measuring the ratio of the absorbances, 595 over 450 nm. This modified Bradford assay is approximately 10 times more than sensitive than the conventional one.[18]
The Coomassie Blue G250 dye used to demark to the proteins in the original Bradford method readily binds to arginine and lysine groups of proteins. This is a disadvantage because the preference of the dye to demark to these amino acids can consequence in a varied response of the assay between different proteins. Changes to the original method, such as increasing the pH past adding NaOH or adding more than dye have been made to correct this variation. Although these modifications result in a less sensitive assay, a modified method becomes sensitive to detergents that can interfere with sample.[nineteen]
Sample Bradford procedure [edit]
Materials [edit]
- Lyophilized bovine plasma gamma globulin
- Coomassie brilliant bluish 1
- 0.xv M NaCl
- Spectrophotometer and cuvettes
- Micropipettes
Process (Standard Assay, 20-150 µg poly peptide; 200-1500 µg/mL) [edit]
- Prepare a series of standards diluted with 0.xv M NaCl to final concentrations of 0 (blank = No protein), 250, 500, 750 and 1500 µg/mL. Also fix series dilutions of the unknown sample to be measured.
- Add 100 µL of each of the to a higher place to a split up test tube (or spectrophotometer tube if using a Spectronic 20).
- Add v.0 mL of Coomassie Bluish to each tube and mix by vortex, or inversion.
- Adjust the spectrophotometer to a wavelength of 595 nm, using the tube which contains no protein (blank).
- Await 5 minutes and read each of the standards and each of the samples at 595 nm wavelength.
- Plot the absorbance of the standards vs. their concentration. Compute the extinction coefficient and summate the concentrations of the unknown samples.
Process (Micro Assay, 1-10 µg protein/mL) [edit]
- Prepare standard concentrations of protein of 1, 5, 7.v and 10 µg/mL. Ready a blank of NaCl merely. Prepare a series of sample dilutions.
- Add 100 µL of each of the above to separate tubes (utilize microcentrifuge tubes) and add 1.0 mL of Coomassie Blue to each tube.
- Turn on and arrange a spectrophotometer to a wavelength of 595 nm, and bare the spectrophotometer using one.five mL cuvettes.
- Wait ii minutes and read the absorbance of each standard and sample at 595 nm.
- Plot the absorbance of the standards vs. their concentration. Compute the extinction coefficient and calculate the concentrations of the unknown samples.
Using data obtained to find concentration of unknown [edit]
In summary, in order to find a standard curve, 1 must use varying concentrations of BSA (Bovine Serum Albumin)[two] in order to create a standard bend with concentration plotted on the ten-axis and absorbance plotted on the y-centrality. Only a narrow concentration of BSA is used (2-10 ug/mL) in guild to create an authentic standard curve.[20] Using a broad range of poly peptide concentration volition make information technology harder to decide the concentration of the unknown protein. This standard curve is then used to make up one's mind the concentration of the unknown poly peptide. The following elaborates on how 1 goes from the standard curve to the concentration of the unknown.
First, add together a line of best fit, or Linear regression and display the equation on the nautical chart. Ideally, the R2 value volition exist as close to 1 every bit possible. R represents the sum of the square values of the fit subtracted from each data point. Therefore, if R2 is much less than one, consider redoing the experiment to get ane with more reliable data.[21]

Graph ane. Actual BSA data attained from a micro calibration UV-Vis Spectrophotometer
The equation displayed on the nautical chart gives a ways for calculating the absorbance and therefore concentration of the unknown samples. In Graph 1, x is concentration and y is absorbance, and so one must rearrange the equation to solve for x and enter the absorbance of the measured unknown.[22] It is likely that the unknown will have absorbance numbers outside the range of the standard. These should non be included calculations, every bit the equation given cannot utilise to numbers exterior of its limitations. In a large scale, one must compute the extinction coefficient using the Beer-Lambert Police force A=εLC in which A is the measured absorbance, ε is the slope of the standard curve, Fifty is the length of the cuvette, and C is the concentration beingness determined.[23] In a micro scale, a cuvette may non be used and therefore ane only has to rearrange to solve for x.

Tabular array 1. Actual assay data for determine concentration of unknown based on line of best fit of the above standard curve
In order to attain a concentration that makes sense with the data, the dilutions, concentrations, and units of the unknown must be normalized (Tabular array 1). To practise this, one must split up concentration past volume of protein in order to normalize concentration and multiply past corporeality diluted to right for whatsoever dilution made in the protein before performing the assay.
Culling assays [edit]
Alternative poly peptide assays include:
- Ultraviolet–visible spectroscopy
- Biuret protein assay
- Lowry protein analysis
- BCA protein analysis
- Amido black protein assay
References [edit]
- ^ a b Ninfa, Alexander J; Ballou, David P; Benore, Marilee (2008). Fundamental Laboratory Approaches for Biochemistry and Biotechnology. Wiley. p. 113.
- ^ a b c d Bradford, Marion (1976). "A Rapid and Sensitive Method for the Quantification of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding" (PDF). Belittling Biochemistry. 72 (1–2): 248–254. doi:ten.1006/abio.1976.9999. PMID 942051 – via Google Scholar.
- ^ "Quick Kickoff TM Bradford Protein Assay" (PDF). www.bio-rad.com.
- ^ Bradford, Marion M. (May 1976). "A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding". Analytical Biochemistry. 72 (1–2): 248–254. doi:10.1006/abio.1976.9999. PMID 942051.
- ^ "Poly peptide conclusion by the Bradford method".
- ^ a b c Kruger, Nicholas J. (2009), Walker, John Grand. (ed.), "The Bradford Method For Protein Quantitation", The Poly peptide Protocols Handbook, Totowa, NJ: Humana Press, pp. 17–24, doi:10.1007/978-1-59745-198-7_4, ISBN978-i-60327-474-6 , retrieved 2022-06-27
- ^ P., Ballou, David; Marilee., Benore (2010). Central laboratory approaches for biochemistry and biotechnology. John Wiley. ISBN9780470087664. OCLC 420027217.
- ^ Ninfa, Ballou, Benore, Alexander J., David P., Marilee (2010). Fundamental Laboratory Approaches for Biochemistry and Biotechnology. The states of America: John Wiley & Sons, Inc. pp. 110, 113. ISBN978-0-470-08766-4.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (2010). Cardinal Laboratory Approaches for Biochemistry and Biotechnology. John Wiley & Sons Inc. p. 113. ISBN978-0470087664.
- ^ a b Okutucu, Burcu; Dınçer, Ayşşe; Habib, Ömer; Zıhnıoglu, Figen (2007-08-01). "Comparison of five methods for decision of full plasma protein concentration". Journal of Biochemical and Biophysical Methods. 70 (5): 709–711. doi:10.1016/j.jbbm.2007.05.009. PMID 17597224.
- ^ "Protein Analysis Technical Handbook" (PDF).
- ^ "4.5. Determination of protein concentration". elte.prompt.hu. Archived from the original on 2016-09-21. Retrieved 2016-05-19 .
- ^ barbosa, Helder; Slater K.H., Nigel (iii August 2009). "Protein quantification in the presence of poly(ethylene glycol) and dextran using the Bradford method". Analytical Biochemistry. 395 (ane): 108–110. doi:10.1016/j.ab.2009.07.045. PMID 19653991.
- ^ Ninfa, Alexander J. (2010). Key Laboratory Approaches for Biochemistry and Biotechnology. Wiley. pp. 117–118. ISBN978-0470087664.
- ^ Ninfa, Ballou (1998). Primal Approaches to Biochemistry and Biotechnology. Fitzgerald Science Press, Bethesda, Medico. pp. 114–116. ISBN978-0470087664.
- ^ Ninfa, Alexander J; Ballou, David P; Benore, Marilee (2009). Fundamental Laboratory Approaches for Biochemistry and Biotechnology. Wiley. p. 113.
- ^ Rabilloud, Thierry (2018). "Optimization of the cydex blue assay: A ane-step colorimetric protein assay using cyclodextrins and compatible with detergents and reducers". PLOS I. 13 (iv): e0195755. Bibcode:2018PLoSO..1395755R. doi:ten.1371/journal.pone.0195755. PMC5895047. PMID 29641569.
- ^ Zor, Tsaffrir; Selinger, Zvi (1996-05-01). "Linearization of the Bradford Protein Assay Increases Its Sensitivity: Theoretical and Experimental Studies". Belittling Biochemistry. 236 (2): 302–308. doi:10.1006/abio.1996.0171. PMID 8660509.
- ^ Kruger, Nicholas J. (2002). "The Bradford Method for Protein Quantitation". Poly peptide Protocols Handbook, the. pp. 15–22. doi:10.1385/one-59259-169-8:fifteen. ISBN1-59259-169-8.
- ^ "Linearization of the Bradford Protein Analysis Increases Its Sensitivity: Theoretical and Experimental Studies" (PDF). www.tau.ac. Nov 20, 1995.
- ^ Albright, Brian (2009). Mathematical Modeling with Excel. p. 60. ISBN978-0763765668.
- ^ Stephenson, Frank Harold (2003). Calculations for molecular biological science and biotechnology: a guide to mathematics in the laboratory . pp. 252. ISBN978-0126657517.
- ^ Ibanez, Jorge G. (2007). Environmental chemistry: fundamentals . pp. lx. ISBN978-0387260617.
Further reading [edit]
- Bradford, Chiliad.M. (1976), "Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of poly peptide-dye binding", Anal. Biochem., 72 (1–two): 248–254, doi:10.1016/0003-2697(76)90527-3, PMID 942051
- Zor, T.; Selinger, Z. (1996), "Linearization of the Bradford poly peptide assay increases its sensitivity: theoretical and experimental studies", Anal. Biochem., 236 (2): 302–308, doi:10.1006/abio.1996.0171, PMID 8660509
- Noble, James E.; Bailey, Marc J.A. (2009). "Chapter 8 Quantitation of Protein". Guide to Protein Purification, 2nd Edition. Methods in Enzymology. Vol. 463. pp. 73–95. doi:ten.1016/S0076-6879(09)63008-1. ISBN9780123745361. PMID 19892168.
- Albright, Brian (2009), Mathematical Modeling with Excel, p. sixty, ISBN978-0763765668
- Stephenson, Frank Harold (2003), Calculations for molecular biology and biotechnology: a guide to mathematics in the laboratory , pp. 252, ISBN978-0126657517
- Dennison, C. (2013). A Guide to Protein Isolation. Springer Science & Business Media. p. 39. ISBN978-94-017-0269-0.
- Ibanez, Jorge 1000. (2007), Environmental chemical science: fundamentals, p. 60, ISBN978-0387260617
External links [edit]
- Bradford assay chemistry
- Variable Pathlength Spectroscopy
- OpenWetWare
Coomassie Protein Assay Reagent Sds,
Source: https://en.wikipedia.org/wiki/Bradford_protein_assay
Posted by: finleysionuirt.blogspot.com


0 Response to "Coomassie Protein Assay Reagent Sds"
Post a Comment